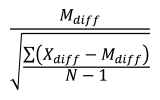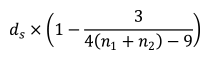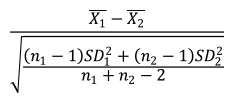Decoded Neurofeedback Project
within Strategic Research Program for Brain Sciences (SRPBS),
BMI Technology Application of DecNef for development of diagnostic and care system for mental disorders and construction of clinical application bases
Consortium
In 2013, around 60 neuroscientists, neuropsychiatrists, engineers, computational scientists, and psychologists from eight Japanese universities and institutes formed a consortium as part of the Japanese Strategic Research Program for the Promotion of Brain Science (SRPBS) for big data applications, machine-learning algorithms, and sophisticated fMRI neurofeedback methods1-3 for the diagnosis and treatment of multiple psychiatric disorders. Many of these techniques are related to brain machine interfaces. SRPBS,4 a nation-wide research program for brain science is supported by the Japanese Advanced Research and Development Programs for Medical Innovation (AMED). Here, we explain the consortium’s organization, its data collection and sharing projects, its sophisticated neurofeedback projects, the construction of biomarkers for multiple psychiatric disorders, therapy based on sophisticated neurofeedback methods, and the biological dimensions for the spectrum of multiple disorders.
Organization chart
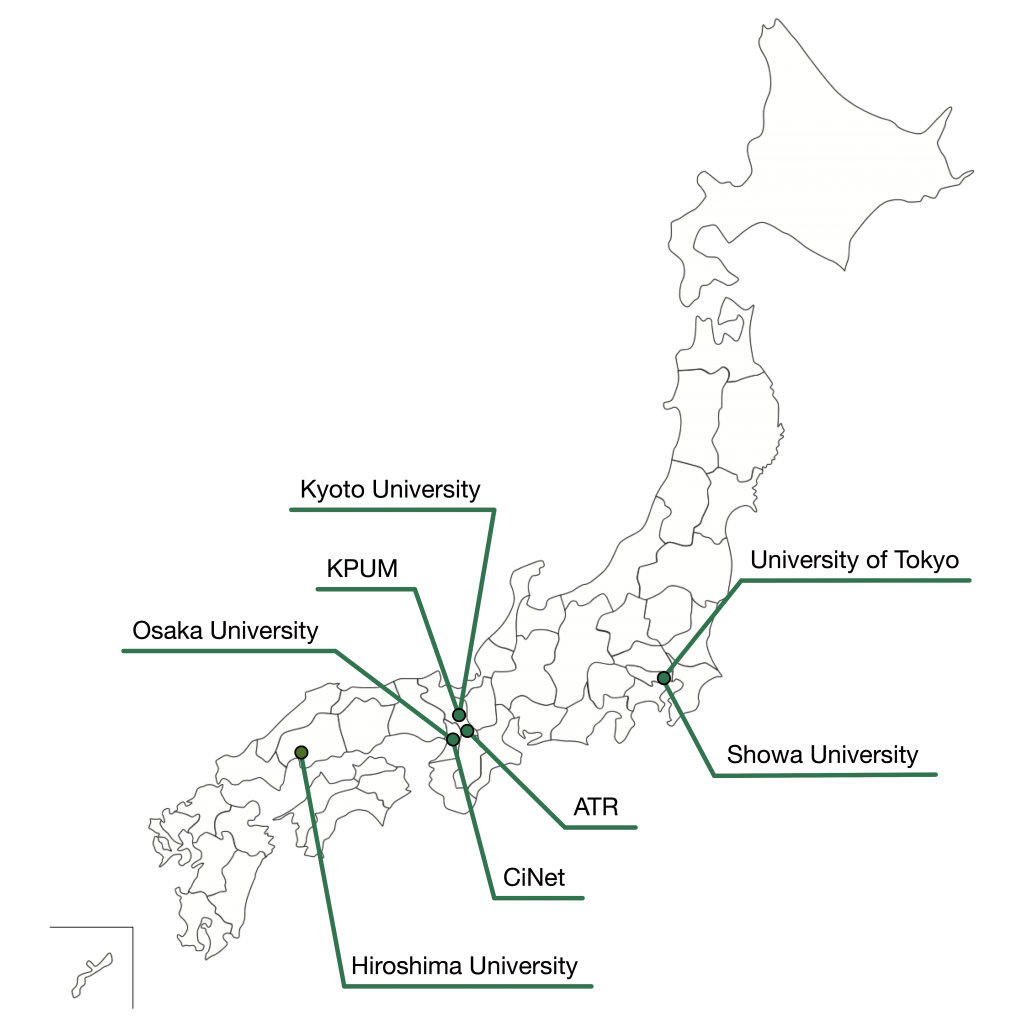
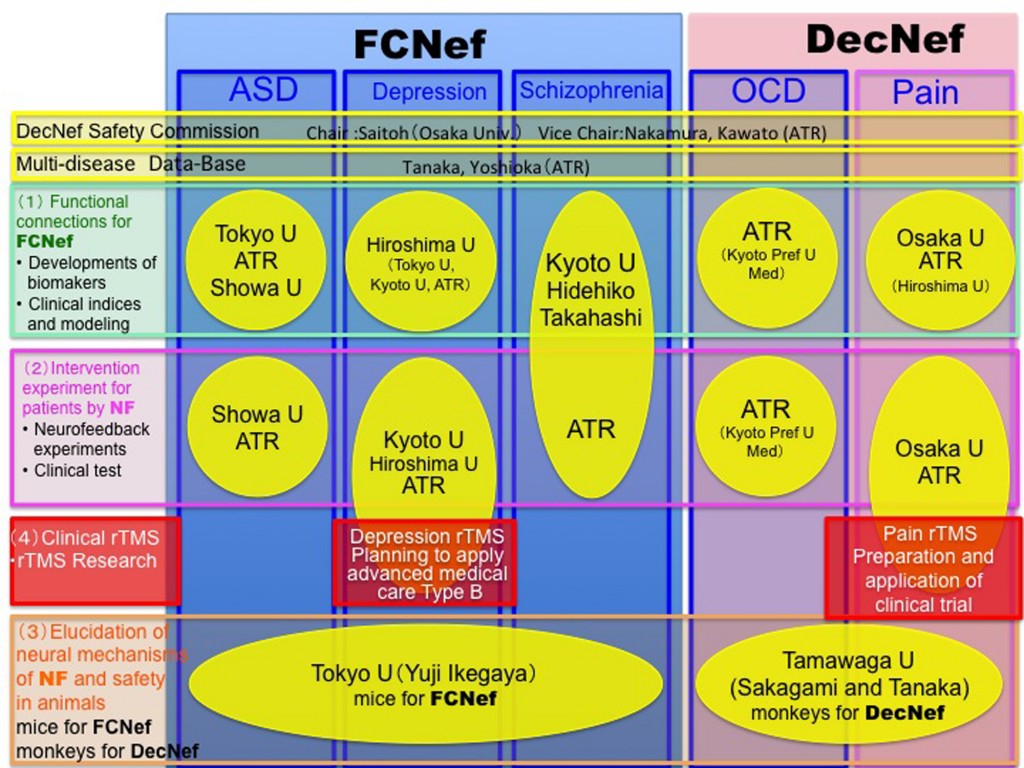
The consortium consists of eight Japanese universities and institutes (Figs. 1 and 2), and approximately 60 researchers participate (see list of participants).
Group Leader: Mitsuo Kawato (ATR Brain Information Communication Research Laboratory Group), members
Sub-Group Leaders:
Noriaki Yahata, Tsuyoshi Araki (Tokyo University), members
Masamichi Sakagami (Tamagawa University), members
Hidehiko Takahashi (Kyoto University), members
Youichi Saitoh (Osaka University), members
Yasumasa Okamoto (Hiroshima University), members
Nobumasa Kato (Showa University), members
From 2013〜 (MEXT and AMED)
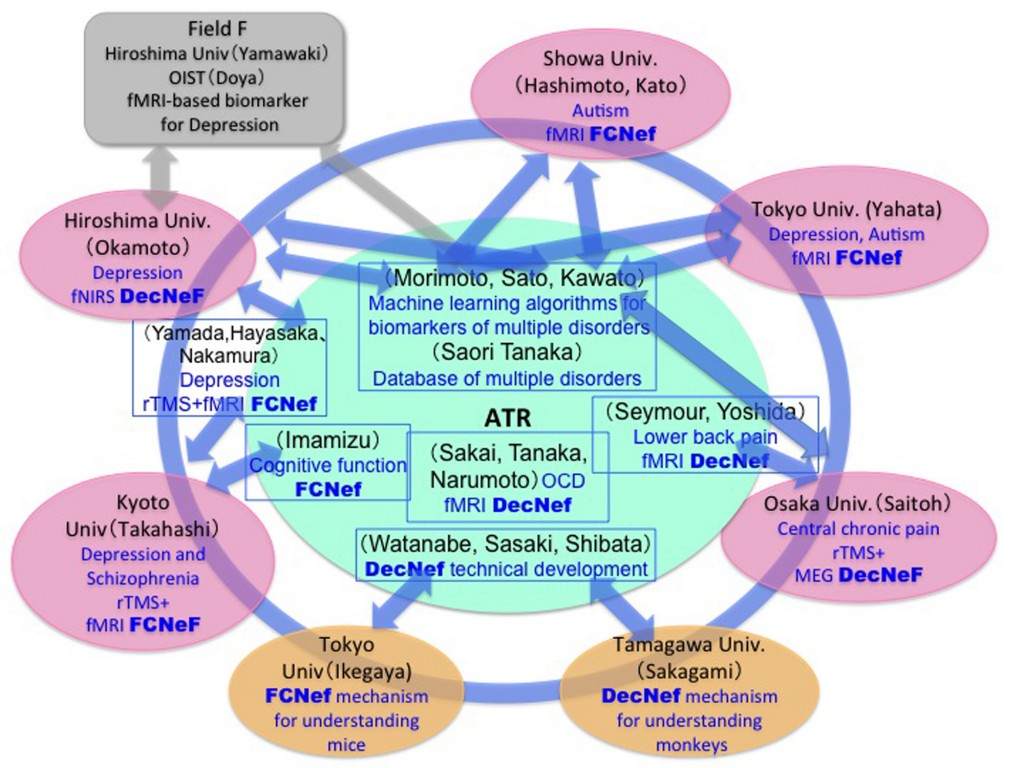
Figure 1
Data collection and sharing
The neuroimaging data of the patients and the control participants were collected at the sites shown in Figure 3. A coherent protocol was designed and used for the resting-state functional magnetic resonance imaging (rs-fMRI). 5 The following table 1 shows the number of samples collected for each disorder at each site. We used altogether 14 scanners from three manufacturers. The data is available from this site. More than 400 scans were obtained for 9 traveling subjects who visited 12 scanners as shown in Table 2.
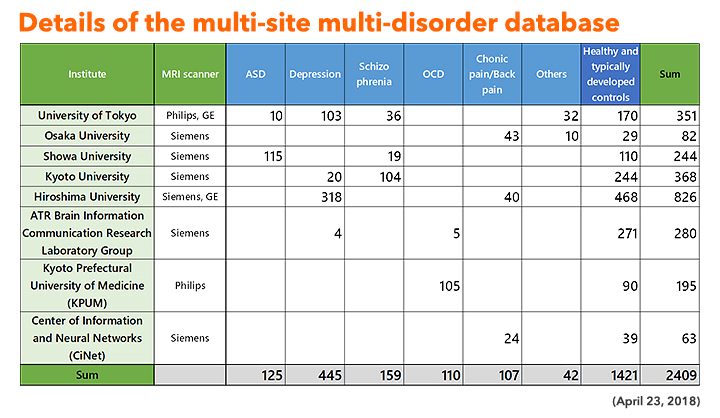 Table 1
Table 1
 Table 2
Table 2
DecNef and FCNef
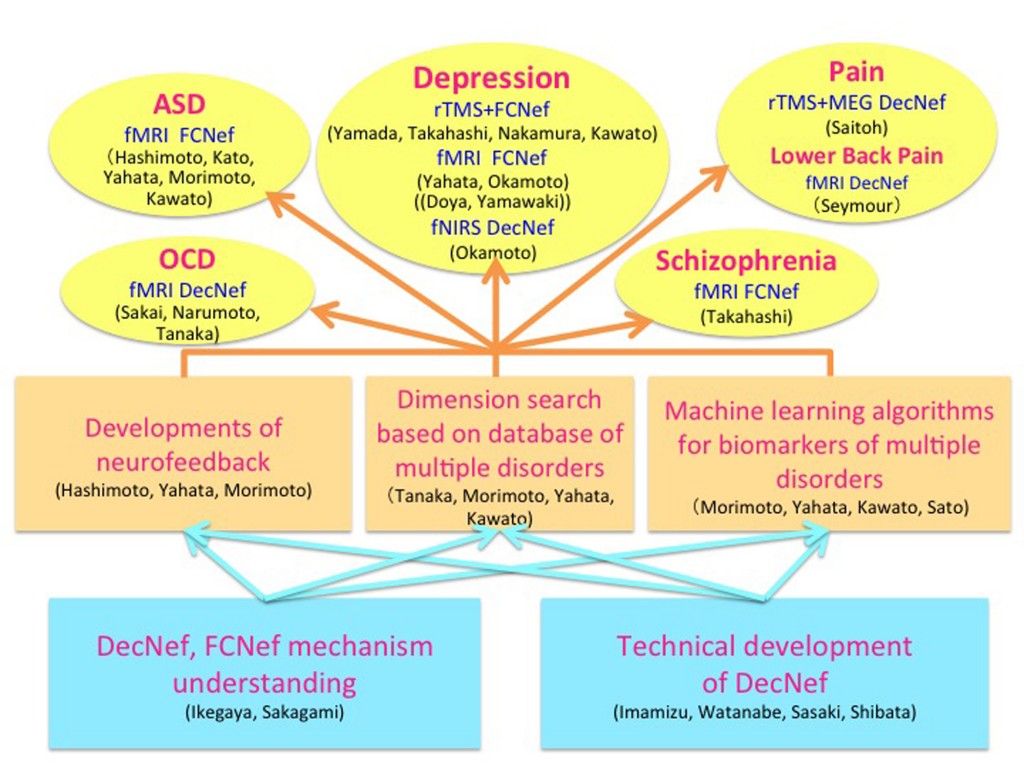
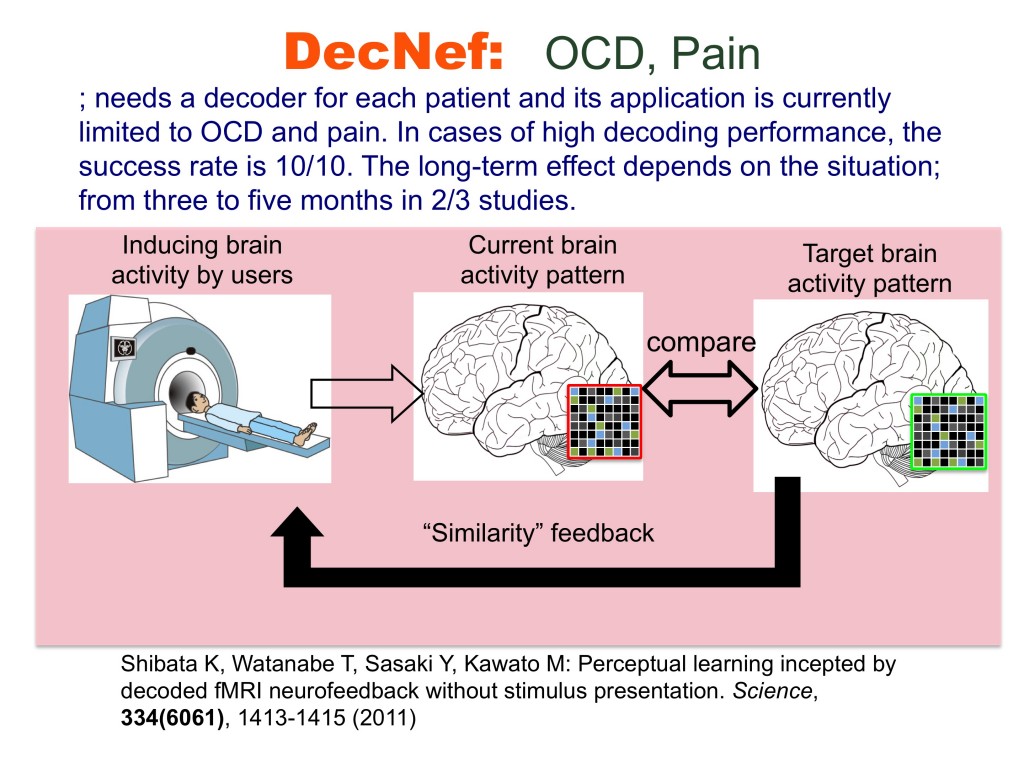
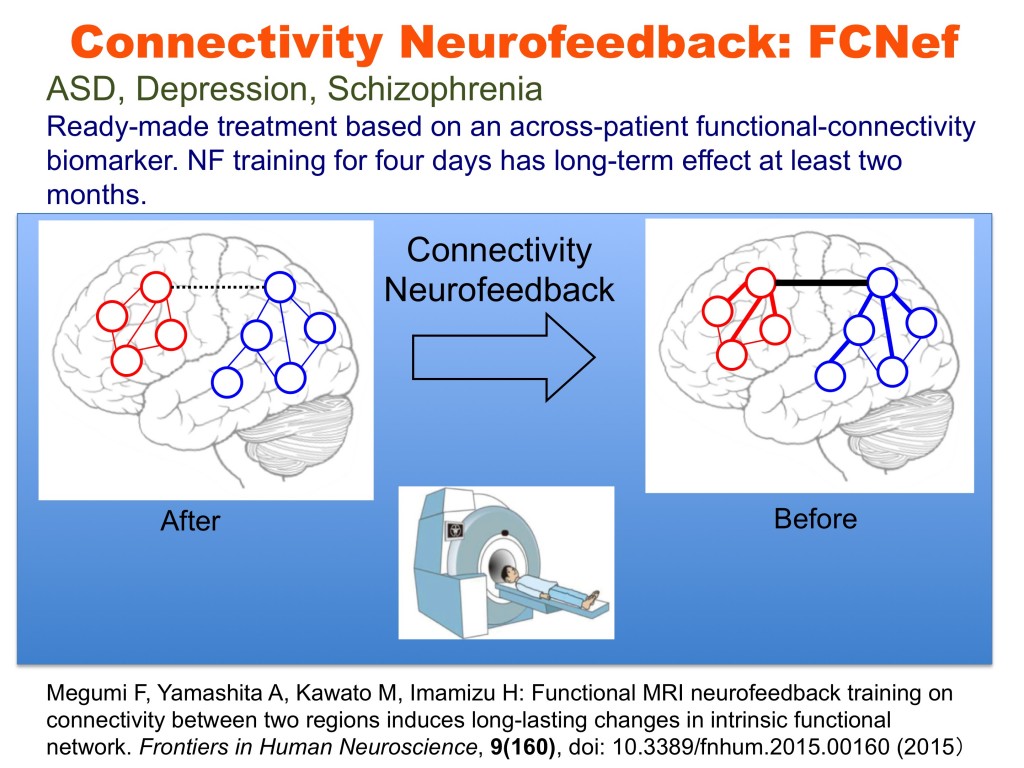
The brain is not merely an input-output information transformation system; it’s also a dynamical system that generates spontaneous spatiotemporal patterns even without sensory inputs, executed movements, or cognitive tasks. These spontaneously generated patterns by brain dynamics are called spontaneous brain activities in experimental animals and resting-state brain activities in humans. The resting-state brain activity of an individual contains rich information about age, cognitive capability, mental disorders, etc. By combining the information decoded from brain activity and its neurofeedback in reinforcement learning paradigms, we can unconsciously control brain activity patterns that correspond to specific information. This leads to therapies for psychiatric disorders3, unconscious manipulation of facial preferences6, color-orientation association7, fear memory reduction8, confidence in decision making9,10, and an increase of cognitive capability. Decoded neurofeedback (DecNef)1 has been applied to obsessive compulsive disorder (OCD) and central chronic pain11 (Fig. 4). Functional connectivity neurofeedback (FCNef)2 has been applied to the autism spectrum disorder (ASD), major depressive disorder (MDD)12, and schizophrenia (SCZ) (Fig. 5), (Table 3).
Effect sizes in DecNef and FCNef studies
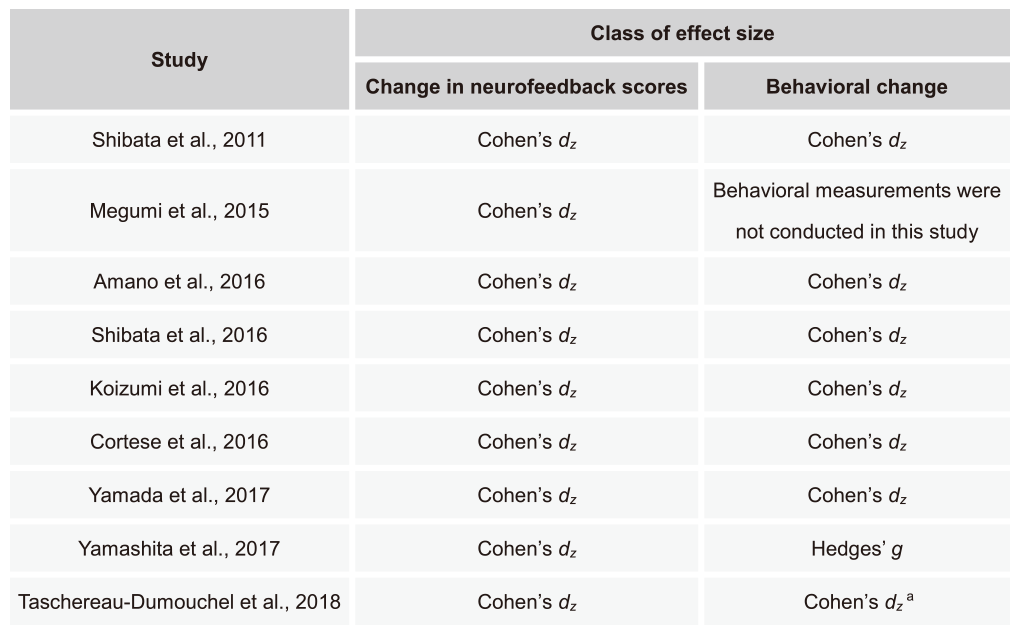
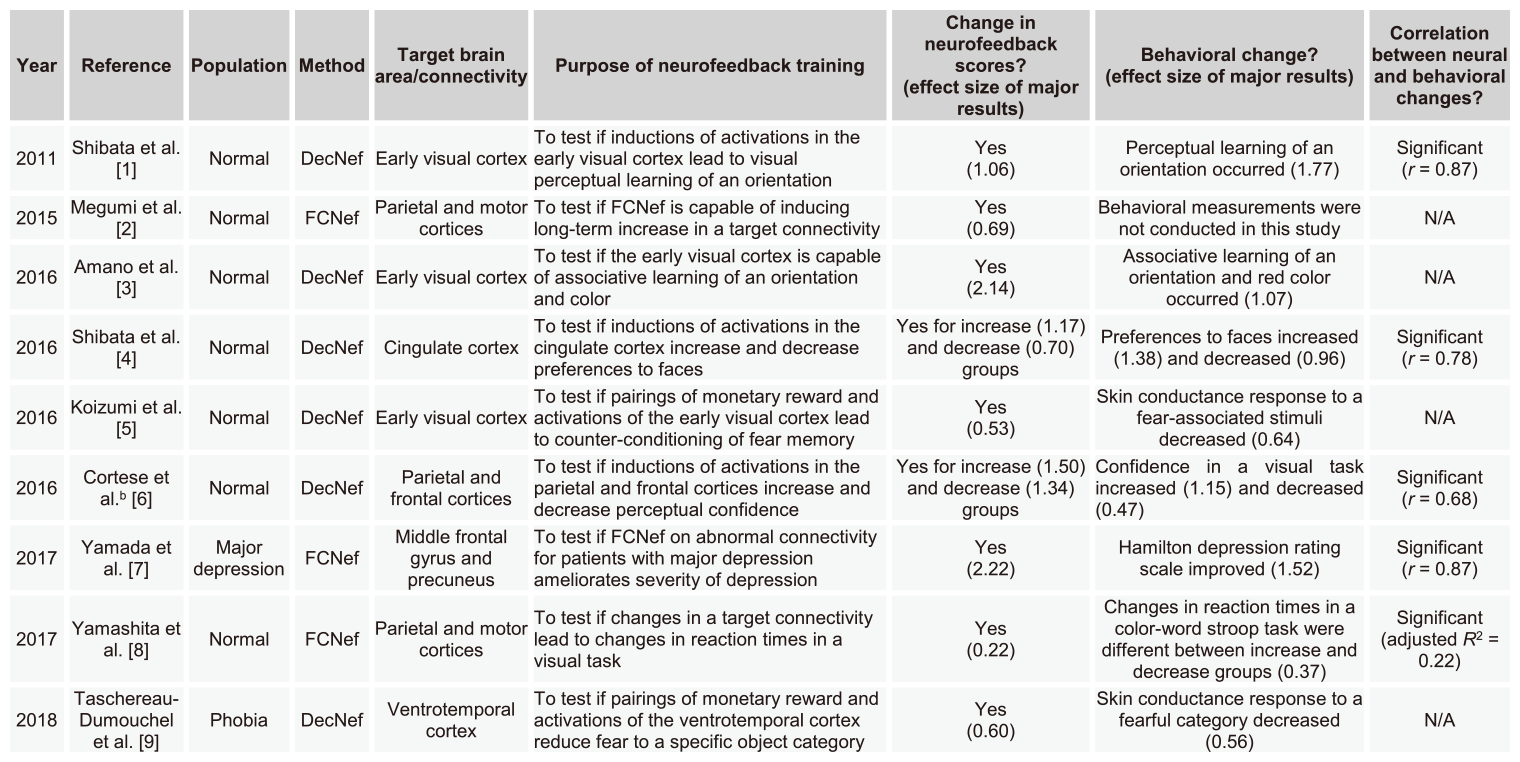
Here, effect sizes in our 9 DecNef and FCNef studies are described. For a review, please see Watanabe et al., Trends in Cognitive Sciences, 2017. Note that in previously published version of datasets in the review Cohen’s d or Cohen’s dz was shown as an effect size of within-subject changes for each study. Here we show only Cohen’s dz. The reason that Hedges’ g is shown for behavioral data in Yamashita et al., Cereb Cortex, 2017 is that the numbers of subjects from the groups compared were not the same.
Calculation of effect sizes
Table 4 (below) summarizes types of effect sizes used for each of the 9 DecNef/FCNef studies shown in Table 5. In the following sections, we show specific formulas for the calculation of effect sizes. Please refer to Lakens, Front Psychol, 2013 for further details of the calculation of the effect sizes.
a The ogirinal paper used Cohen’s d (corrected for dependence between the means). Here Cohen’s dz is used to preserve consistency with other studies shown on this table.
Table 4
Table 4. Methods for calculation of effect sizes in each study. Each row corresponds to each study using DecNef or FCNef shown in Table 5. The left column shows the name of the first author and published year for each study. The right 2 columns indicate types of effect sizes for changes in neurofeedback scores and behaviors.
Cohen’s dz was defined as:
where the numerator represents the difference between the mean (Mdiff) of the difference scores (Xdiff) and the comparison value (e.g., 0), and the denominator represents the standard deviation of the difference scores. N indicates the number of samples.
Hedges’ g was defined as:
where n1 and n2 represent the numbers of samples for 2 groups, respectively. ds is a type of Cohen’s d, defined as:
In this formula, the numerator represents the difference between mean of the 2 groups of observations. The denominator represents the pooled standard deviation.
Raw datasets used for the calculation of effect sizes
From the following button, one can download an .zip file that contains raw numbers used for the calculation of effect sizes for each of 9 DecNef/FCNef studies.
| This work is licensed under a Creative Commons Attribution 2.5 License |
Table 5. Development and applications of DecNef and FCNef in chronological order.
a N/A, not available
b The authors published another paper (Cortese et al., NeuroImage, 2017) by a different data analysis with a different purpose.
Table 5
- Shibata, K. et al. (2011) Perceptual learning incepted by decoded fMRI neurofeedback without stimulus presentation. Science 334, 1413–1415
- Megumi, F. et al. (2015) Functional MRI neurofeedback training on connectivity between two regions induces long-lasting changes in intrinsic functional network. Front. Hum. Neurosci. 9, 160
- Amano, K. et al. (2016) Learning to associate orientation with color in early visual areas by associative decoded fMRI neurofeedback. Curr. Biol. 26, 1861–1866
- Shibata, K. et al. (2016) Differential activation patterns in the same brain region led to opposite emotional states. PLoS Biol. 14, e1002546
- Koizumi, A. et al. (2016) Fear reduction without fear through reinforcement of neural activity that bypasses conscious expo- sure. Nat. Hum. Behav. 1, 0006
- Cortese, A. et al. (2016) Multivoxel neurofeedback selectively modulates confidence without changing perceptual performance. Nat. Commun. 7, 13669
- Yamada, T. et al. (2017) Resting-state functional connectivity- based biomarkers and functional MRI-based neurofeedback for psychiatric disorders: a challenge for developing theranostic biomarkers. Int. J. Neuropsychopharmacol. 20, 769–781
- Yamashita, A. et al. (2017) Connectivity neurofeedback training can differentially change functional connectivity and cognitive performance. Cereb. Cortex. 27, 4960–4970
- Tascereau-Dumouchel, V. et al. (2018) Towards an unconscious neural-reinforcement intervention for common fears. Proc Nat Acad Sci USA. 115, 3470-3475
How to download data from the DecNef Data Repository (DecNef DR)
We invite investigators to apply for access to the DecNef DR. Before sending your application, please read the privacy statement. The use of data from the DecNef DR is allowed for noncommercial purpose only. Please note that we collect the number of applicants and size of the data downloaded from the database for reports to our funding agencies (ImPACT, ERATO, etc), without private information.
We kindly ask you to email us the following form of “Declaration of Consent”.
- E-mail : open-decnef__AT__atr.jp (Please replace __AT__ with @.)
- Declaration of Consent : PDF
You will be notified by email how to access the database, as well as where you may log in and download data to your computer. If you have any questions about the data or the database, please contact us by email.
NOTE: i) everyone who works with the DecNef DR must review and agree to the terms of use, including those who are accessing shared copies of the data. If you are sharing data from the DecNef DR, please let your collaborators know that they must register and agree to the terms of use.
How to cite the use of data from the DecNef DR
We ask that you kindly acknowledge the source of the data in your published manuscripts, conference presentations, etc as follows:
Data collection and sharing for this project was provided by the DecNef Department at the Advanced Telecommunication Research Institute International, Kyoto, Japan.
Biomarkers of ASD, SCZ, MDD and OCD based on rs-fcMRI
Recently, using functional-connectivity magnetic resonance imaging (fcMRI) techniques, attempts have developed classifiers for various psychiatric disorders and typically developed or healthy individuals to identify the abnormality of functional connections (FCs). Because no classifiers have ever been validated for a completely independent cohort due to the over-fitting and interferential effects of varying measurement conditions and demographic distributions, their clinical utility and neuroscientific validity continue to be questioned. Within this consortium, we overcame the above difficulties by developing a novel machine-learning algorithm that identifies a small number of abnormal FCs.12-14 The resultant classifier achieved high accuracy for a Japanese discovery cohort and also demonstrated good generalizations for independent validation cohorts collected in the USA and/or Netherlands. The combination of L1-SCCA and SLR has already been applied to the classification of ASD13, SCZ15, MDD16, and OCD17 patients from typically developed or healthy controls. Table 6 shows the results of these studies as of December 2017. The classifiers performances continue to improve.
DecNef and FCNef therapy
Neurofeedback training based on real-time fMRIs has attracted considerable attention for its potential advantages in therapeutic treatment. Most fMRI neurofeedback methods focus on the up- or down-regulation of single region-of-interest activation, but they have recently been extended by DecNef to control spatial activity patterns within a specific ROI. DecNef can be used as a noninvasive BMI system for treating abnormal central decision-making functions in psychiatric disorders. Megumi Fukuda, Ayumu Yamashita, Kawato and Imamizu2,18 also demonstrated that fMRI neurofeedback can further modulate spatiotemporal activation patterns across multiple ROIs as well as intrinsic networks. This effect was preserved over a surprisingly long period. Disturbances in intrinsic networks have been reported for a number of neurological and psychiatric diseases. Consequently, the connectivity neurofeedback method could be applied not only as a causal tool for basic neuroscience to alter functional networks but also as a revolutionary therapeutic method by directly curing the pathological connectivity patterns of neurological and psychiatric diseases3,12,14. DecNef or FCNef may provide revolutionary cures for psychiatric diseases.
The utility of our developed rs-fcMRI classifier is found in its neurofeedback therapy, especially FCNef.2,18 Megumi et al. (2015)2 found that four days of fMRI real-time neurofeedback of functional connectivity between two designated areas persistently changed the rs-fcMRI connectivity for more than two months. Within the above SRPBS consortium, we previously applied this connectivity fMRI neurofeedback method to ASD individuals with some preliminarily encouraging results12. The target FCs were selected by the ASD classifier in the following ways.12-14 The WLS of the ASD biomarker was estimated in a real-time fMRI neurofeedback paradigm for an ASD patients, and a sign-inverted WLS value was fed back to the ASD patient as a monetary reward in a reinforcement learning framework. We expect increases in neurofeedback scores, in other words, reductions in the WLS values, to lead to attaining normal network dynamics in rs-fcMRIs.
Decoded neurofeedback (DecNef)1 has been applied to obsessive compulsive disorder (OCD) and central chronic pain, and preliminarily promising results were obtained.3,11 DecNef was especially applied to 15 patients with pain and induced statistically significant decreases in VAS compared with an adequate sham feedback control. Functional connectivity neurofeedback (FCNef)2 was applied to autism spectrum disorder (ASD), major depressive disorder (MDD), and schizophrenia (SCZ) with preliminarily promising results as well as over 15 ASD patients. Improvements of several clinical indices were observed. For a few cases, even two weeks after FCNef intervention, the resting-state intrinsic network remained in a normal pattern. 12
Spectrum of Psychiatric Disorders revealed by Artificial Intelligence and Big Data
The current diagnosis and treatment of psychiatric disorders with childhood and adult onsets are categorical, as described by DSM-5. However, a large GWAS study identified the genetic risk loci shared by several disorders, and a meta-analysis found decreases common to several disorders in the gray matter volumes of specific brain regions; therefore, more emphasis has recently been placed on the spectrum of disorders and the exploration of biological dimensions to characterize it. This line of research might provide a neuroscience background and explain why a single drug is efficient for multiple disorders. We formed a consortium (SRPBS-DecNef) in 2013 to develop diagnosis and therapy based on computational neuroscience and machine-learning algorithms. By utilizing resting-state functional connectivity MRI (rs-fcMRI), we developed biomarkers of autism spectrum disorder (ASD), schizophrenia, major depressive disorder (MDD), and obsessive-compulsive disorder (OCD). More than 2,000 samples were collected from more than ten scanners. Our aim is to develop new therapies using decoded fMRI real-time neurofeedback (DecNef), based on multi-voxel pattern analysis,1 or connectivity real-time neurofeedback (FCNef) based on rs-fcMRI biomarkers.2 Here, we introduce a few studies that support a spectrum of psychiatric disorders.
Yahata et al. (2016)13 successfully developed an ASD classifier with 85% accuracy for three Japanese sites and a marked generalization capability with 75% accuracy for the USA ABIDE dataset.13 A machine-learning algorithm, which consisted of a nested feature selection by L1-SCCA and cross validation by sparse logistic regression, only selected 16 functional connections (FCs) from 9,730 FCs among 140 brain regions. The same algorithm was also applied to schizophrenia, MDD, and OCD with comparable performances (Table 6, and the computer code is available here). The four classifiers selected about 60 FCs, and 1,000 samples were hierarchically grouped within these 60-FC coordinates into five clusters. The hierarchical structure turned out to be ((((ASD, healthy), SCZ), OCD), MDD), and each cluster corresponded to diagnostic labels with 51–85% accuracy; the overall distribution of the samples formed a spectrum with significant overlaps among disorders.
Yamashita et al. (2015)19 predicted the learning plateau of a verbal 3-back working memory task in 17 healthy young Japanese participants by a linear weighted summation of 16 FCs among 18 functional brain networks. Their model explained the individual working memory capabilities of a much larger and more greatly diversified American dataset. It also predicted the individual differences in the working memory ability in schizophrenic patients of the SRPBS-DecNef consortium. The severity of the working memory deficit is in the order of schizophrenia, MDD, OCD, and ASD. The model reproduced not only this order but also the quantitative aspects of the deficits in different psychiatric disorders. If the neural mechanisms connecting functional brain networks with cognitive (dys) functions in different psychiatric disorders were different, no model, which was developed using only healthy participants, could have been generalized to multiple disorders. Therefore, the spectrum view of psychiatric disorders is supported at the level that connects macro-functional connectomes with cognitive functions. A common neural mechanism seems to connect functional networks with syndromes, and only FCs might be characteristically different among various psychiatric disorders.
- Shibata K, Watanabe T, Sasaki Y, and Kawato M: Perceptual learning incepted by decoded fMRI neurofeedback without stimulus presentation, Science, 334, 1413-1415 (2011)
- Megumi F, Yamashita A, Kawato M, and Imamizu H: Functional MRI neurofeedback training on connectivity between two regions induces long-lasting changes in intrinsic functional network. Front Hum Neurosci 9, 160 (2015).
- Watanabe T, Sasaki Y, Shibata K, Kawato M: Advances in fMRI real-time neurofeedback, Trends in Cognitive Sciences, 21(12), 997-1010 (2017) .
- Home page for SRPBS http://www.nips.ac.jp/srpbs/index.html
- ATR protocol page https://bicr.atr.jp/rs-fmri-protocol-2/
- Shibata K, Watanabe T, Kawato M, Sasaki Y: Differential activation patterns in the same brain region led to opposite emotional states, PLoS Biology, 14(9): e1002546 (2016)
- Amano K, Shibata K, Kawato M, Sasaki Y, Watanabe T: Learning to associate orientation with color in early visual areas by associative decoded fMRI neurofeedback, Curr Biol, 26(14), 1861–1866 (2016)
- Koizumi A, Amano K, Cortese A, Shibata K, Yoshida W, Seymour B, Kawato M, Lau H: Fear reduction without fear through reinforcement of neural activity that bypasses conscious exposure, Nature Human Behavior, 1:0006, doi:10.1038/s41562-016-0006 (2016)
- Cortese A, Amano K, Koizumi A, Kawato M, Lau H: Multivoxel neurofeedback selectively modulates confidence without changing perceptual performance, Nature Communications, 7:13669 (2016)
- Cortese A, Amano K, Koizumi A, Lau H, Kawato M: Decoded fMRI neurofeedback can induce bidirectional confidence changes within single participants, NeuroImage, 149, 323–337 (2017)
- Yanagisawa T, Fukuma R, Seymour B, Hosomi K, Kishima H, Shimizu T, Yokoi H, Hirata M, Yoshimine T, Kamitani Y, Saitoh Y: Induced sensorimotor brain plasticity controls pain in phantom limb patients. Nature Communications. 7:13209. doi: 10.1038/ncomms13209. (2016)
- Yamada T, Hashimoto R, Yahata N, Ichikawa N, Yoshihara Y, Okamoto Y, Kato N, Takahashi H, Kawato M: Resting-state functional connectivity-based biomarkers and functional MRI-based neurofeedback for psychiatric disorders: a challenge for developing theranostic biomarkers, Int J Neuropsychopharmacol, 20(10): 769–781, (2017) .
- Yahata N, Morimoto J, Hashimoto R, Lisi G, Shibata K, Kawakubo Y, Kuwabara H, Kuroda M, Yamada T, Megumi F, Imamizu F, Náñez Sr JE, Takahashi H, Okamoto Y, Kasai K, Kato K, Sasaki Y, Watanabe T, and Kawato M: A Small Number of Abnormal Brain Connections Predicts Adult Autism Spectrum Disorder. Nature Communications. 7:11254. doi: 10.1038/ncomms11254 (2016).
- Yahata N, Kasai K, Kawato M: Computational neuroscience approach to biomarkers and treatments for mental disorders, Psychiatry and Clinical Neurosciences, 71: 215–237 (2017)
- Yoshihara N, Lisi G, Yahata N, Fujino J, Matsumoto Y, Miyata J, Sugihara G, Urayama S, Kubota M, Yamashita M, Hashimoto R, Ichikawa N, van Haren NEM, Mori S, Okamoto Y, Kasai K, Kato N, Imamizu H, Kahn RS, Sawa A, Kawato M, Murai T, Morimoto J, Takahashi H:Schizophrenia biomarker using whole brain resting-state functional connectivity MRI: generalization to independent cohorts from different countries and disorder stages, Society for Neuroscience 47th Annual Meeting (2017).
- Ichikawa N, Lisi G, Yahata N, Okada G, Takamura M, Yamada M, Suhara T, Hashimoto R, Yamada T, Yoshihara Y, Takahashi H, Kasai K, Kato N, Yamawaki S, Kawato M, Morimoto J, Okamoto Y: Identifying melancholic depression biomarker using whole-brain functional connectivity, arXiv.org, 1704.01039 (2017)
- Takagi Y, Sakai Y, Lisi G, Yahata N, Abe Y, Nishida S, Nakamae T, Morimoto J, Kawato M, Narumoto J, Tanaka SC: A neural marker of obsessive-compulsive disorder from whole-brain functional connectivity, Scientific Reports, 7(7538) (2017)
- Yamashita A, Hayasaka S, Kawato M, Imamizu H: Connectivity neurofeedback training can differentially change functional connectivity and cognitive performance, Cerebral Cortex, 27;4960–4970, (2017)
- Yamashita M, Kawato M, and Imamizu H: Predicting learning plateau of working memory from whole-brain intrinsic network connectivity patterns, Scientific Reports, 5, doi: 10.1038/srep07622 (2015)
This research was supported by the SRPBS from the Japan Agency for Medical Research and Development, AMED.
created on December 1st, 2015
updated on August 1st, 2016
updated on December 27, 2017